The SARS-CoV-2 (COVID-19) pandemic has shown how ill-prepared modern bioanalytical assays are in combating deadly diseases. This is, in part, due to the major bottlenecks existing in univariant “gold standard” bioanalytical techniques (RT-PCR, microarray, sequencing, ELISA, etc.), which almost exclusively operate in a discrete format unable to assay different classes of biomolecules using single instrument.
LSPR & Biosensing *
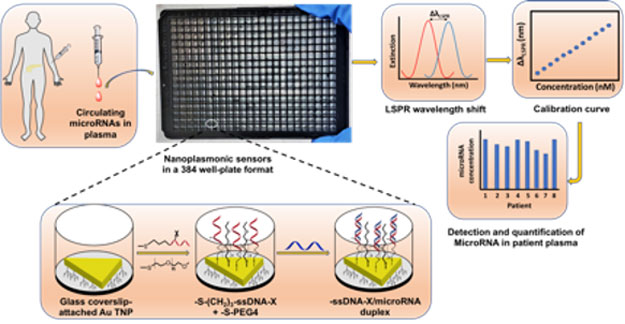
For example, RT-PCR is capable of assaying nucleic acids but not proteins, whereas ELISA only assays proteins but not RNAs. ELISA also shows inadequacies in sensitivity to low abundance biomarkers, resulting in low accuracy. Moreover, PCR and ELISA require large amounts (>200 µL) of patient biofluids, making multiple analyses of samples challenging. In contrast, a technique with the ability to measure multiple classes biomarkers for a disease using a single instrument potentially mitigates false responses, increasing the reliability of the assay. To overcome current challenges in bioanalytical assays and create more accurate, reliable, and reproducible early disease diagnostics, we have fabricated and characterized an entirely new, solid-state, and highly adaptable plasmonic nanosensor (assay switching protocol) capable of measuring microRNAs and proteins from cancer patient plasma and urine samples, respectively, at femtomolar (fM, 10-15 M) sensitivity using a single UV-visible spectrophotometer without any sample processing. We have also designed, fabricated and utilized high-throughput plasmonic nanosensors, which can assay hundreds of samples detecting and quantifying patient plasma-derived microRNAs in a single instrument run. The current research focus of our group is to construct ultrasensitive (attomolar, 10-18 M) highly adaptable plasmonic nanosensors which can detect and quantify multiple microRNAs and proteins simultaneously from in vitro three-dimensional organoid media, and in vivo pancreatic cancer mouse model, and finally a large cohort of gastrointestinal (such as stomach, liver, pancreas, and colorectal) cancer patient plasma specimens. We plan to study the structural parameters of ligands to strongly influence the plasmon electron wavefunction delocalization phenomena and provide a larger LSPR responses, thus enhancing the biosensing sensitivity.
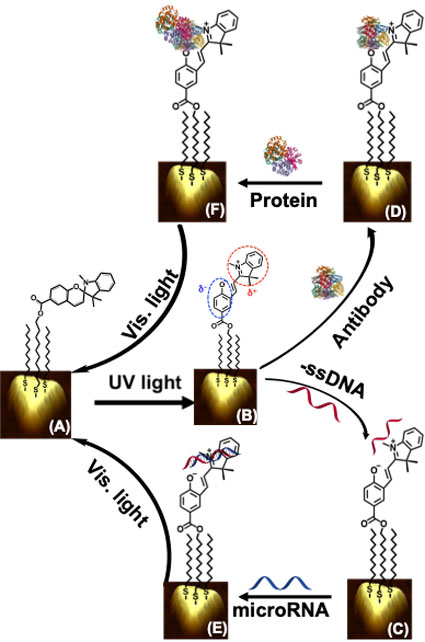
The overarching goal is to conduct multiplexing (assaying multiple biomarkers of a same class, or those with different expression levels such as oncogenic and tumor suppressor microRNAs or three different classes in a single instrument run), and high-throughput (ability to assay at least >15 biomarkers from >20 patient samples simultaneously) nanosensors, which will substantially improve high accuracy disease diagnosis at the clinical setting. We are also conducting a population study by analyzing ~1200 pancreatic disease-related (pancreatic cancer, chronic pancreatitis, and intraductal papillary, mucinous neoplasms) patient samples while detecting ~15 microRNAs and ~10 proteins to develop a more accurate panel of biomarkers for earlier detection of pancreatic cancer, which so far has no cure. We have been collaborating with several world-renowned cancer researchers in Indiana University School of Medicine.
* Collaborators
- Professor Melissa Fisher, Indiana University School of Medicine
- Professor C. Max Schmidt, Indiana University School of Medicine
- Professor Jianjun Zhang, Indiana University School of Medicine
- Professor Hristos Kaimakliotis, Indiana University School of Medicine
- Professor Sha Cao, Indiana University School of Medicine
Representative Publications
- Masterson, A.; Chowdhury, N.; Yang, F.; Yip-Schneider, M.; Hati, S.; Gupta, P.; Cao, S.; Wu, H.; Schmidt, C. M.; Fishel, M.; Sardar, R. Amplification-Free, High-throughput Nanoplasmonic Quantification of Circulating microRNAs in Unprocessed Plasma Microsamples for Earlier Pancreatic Cancer Detection. ACS Sens. 2023, 8, 1085-1100.
- Hati, S.; Langlais, S. R.; Masterson, A. N.; Liyanage, T.; Muhoberac, B. B.; Kaimakliotis, Johnson, M.; Sardar, R. Photoswitchable machine-engineered plasmonic nanosystem with high optical response for ultrasensitive detection of microRNAs and proteins adaptively. (S.H. and S.R.L. contributed equally to this work). Anal. Chem. 2021, 93, 13935-13944.
- Masterson, A. N.; Liyanage, T.; Kaimakliotis, H.; Derami, H. G.; Deiss, F.; Sardar, R. Bottom-up fabrication of plasmonic nanoantenna-based high-throughput multiplexing biosensors for ultrasensitive detection of microRNAs directly from cancer patients’ plasma. Anal. Chem. 2020. 92, 9295-9304.
- Liyanage, T.; Masterson, A. N.; Oyem, H. H.; Kaimakliotis, H.;Nguyen, H.; Sardar, R. Plasmoelectronic-based ultrasensitive assay of tumor suppressor microRNAs directly in patient plasma: Design of highly specific early cancer diagnostic technology. Anal. Chem.2019, 91, 1894-1903.
- Joshi, G. K.; Deitz-McElyea, S.; Liyanage, T.; Lawrence, K. N.; Mali, S.; Sardar, R.; Korc, M. Label-free nanoplasmonic-based short noncoding RNA sensing at attomolar concentration allows for quantitative assay of microRNA-10b in biological fluids and circulating exosomes. ACS Nano. 2015, 9, 11075-11089.
- Joshi, G. K.; Deitz-McElyea, S.; Johnson, M. A.; Mali, S.; *Korc, M.; Sardar, R. Highly specific plasmonic biosensors for ultrasensitive microRNA detection in plasma from pancreatic cancer patients. Nano Lett.2014, 14, 6955-6963.
- Joshi, G. K.; McClory, P.; Dolai, S.; Sardar, R. Improved localized surface plasmon resonance biosensing sensitivity using chemically synthesized gold nanoprisms as plasmonic transducers. Mater. Chem. 2012, 22, 923-931.

